The Missing Semester: Containerization
Building reproducible bioinformatics workflows with Docker.
Welcome to Lesson 1 in Module 3: Scale of Bioinformatics: The Missing Semester!
Introduction
Thus far, we’ve been learning as isolated bioinformaticians. In Module 1, we learned how to conduct boilerplate analyses on three different data types: single cell transcriptomics, spatial transcriptomics and proteomics. In Module 2, we saw how to integrate these different modalities to yield a more holistic picture of biology.
Importantly, our learning has assumed we’re the only ones running said analyses and that we’d only run them a handful of times.
In commercial R&D, this is rarely the case. Bioinformaticians and computational biologists are organized into teams: often interfacing with other disciplines like software or data engineering, wet lab scientists and business stakeholders who guide scientific exploration.
This means that analyses are run far more than a handful of times, on thousands—sometimes millions—of samples. And you aren’t usually the only person running them.
Which brings us to: reproducibility. How do we ensure that when someone else runs our code, it both a) works and b) is shareable?
The answer lies in containers.
A container is a standard unit of software that packages up code and all its dependencies so the application runs quickly and reliably from one computing environment to another (Source).
Perhaps the most commonly used software for containerization—and what we’ll be using from here on out—is Docker.
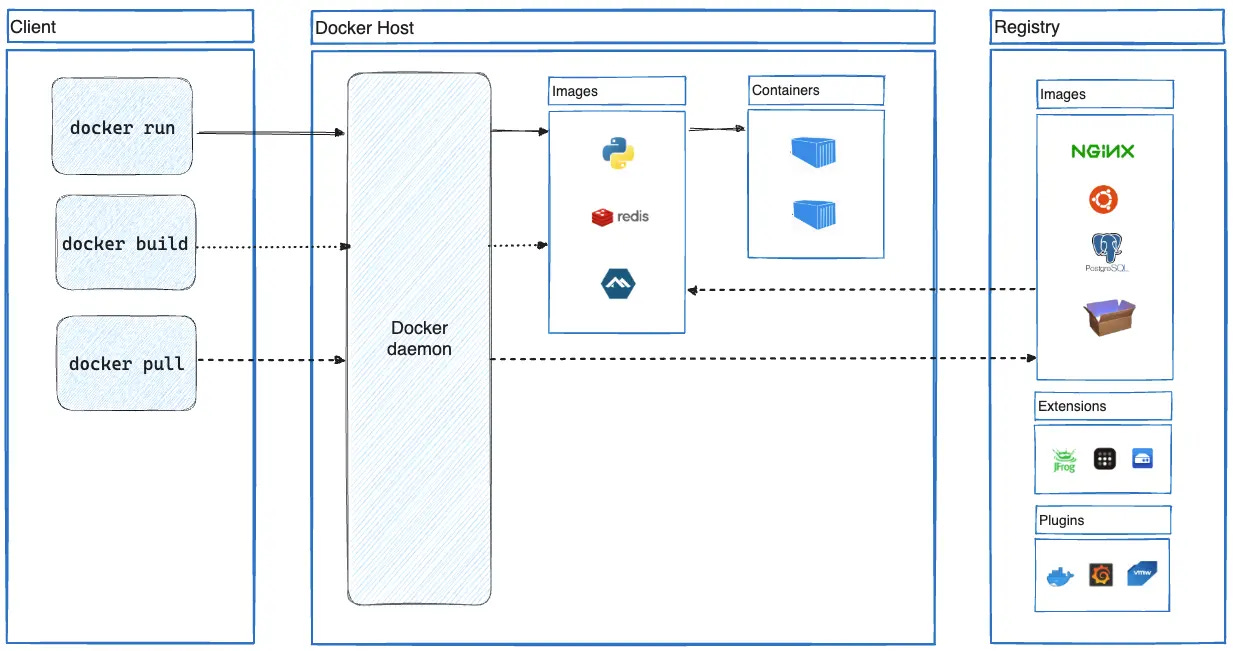
In particular, a Docker container image is a lightweight, standalone, executable package of software that includes everything needed to run an application: code, runtime, system tools, system libraries and settings.
In this lesson, we’ll:
Package a simple script in a Docker container
Run it independently
Walk through sharing our image with colleagues
Before we start
I like to use Docker as a VSC integration for ease (i.e., no switching back and forth between VSC and Docker Desktop). It’s also super intuitive and documentation for getting started with VSC x Docker are plentiful. Before we begin, ensure that you’ve got the following set up:
IDE of choice + Python install
Docker (Docker Desktop for build monitoring and VSC integration for coding)
Write QC script
Imagine we’ve got a team of bioinformaticians that need to annotate their single-cell data with QC metrics for downstream filtering. Let’s write a simple script to do just that:
#!/usr/bin/env python3
import argparse
import scanpy as sc
import anndata as ad
def main(input_file, output_file):
adata = sc.read_h5ad(input_file)
adata.var["mt"] = adata.var_names.str.startswith("MT-")
sc.pp.calculate_qc_metrics(
adata,
qc_vars=["mt"],
percent_top=None,
log1p=False,
inplace=True
)
adata.write(output_file)
print(f"QC metrics added. Output saved to: {output_file}")
if __name__ == "__main__":
parser = argparse.ArgumentParser(description="Annotate MT genes & compute QC metrics on AnnData")
parser.add_argument("input", help="Path to input AnnData .h5ad file")
parser.add_argument("output", help="Path to output AnnData .h5ad file")
args = parser.parse_args()
main(args.input, args.output)This .py script:
Takes an input .h5ad at a specified path
Annotates MT genes
Calculates QC metrics
Writes the updated .h5ad to a new path
We will containerize this script with Docker.
Create Dockerfile
A Dockerfile is it’s own special plain-text format; it isn’t a Markdown, Python or YAML. It’s a set of “instructions” that will tell our application (.py script) how to run.
If you’re working in VSC (this should mostly hold true for PyCharm as well), click “File” in the top-left application pane and in the drop-down, click “New Text File.” When the new file opens up in VSC, paste the following:
# python image
FROM python:3.13-bullseye
# python image
FROM python:3.13-bullseye
# install system dependencies
RUN apt-get update && apt-get install -y \
build-essential \
apt-utils \
ca-certificates \
&& rm -rf /var/lib/apt/lists/*
# set working directory
WORKDIR /app
# copy relevant files into container
COPY qc-metrics.py /app/qc-metrics.py
COPY module3_reqs.txt /app/module3_reqs.txt
# pull dependencies from reqs txt file
RUN pip install uv && uv pip install --system -r module3_reqs.txt
# define container behavior
# always run qc-metrics.py on container startup
ENTRYPOINT ["python", "/app/qc-metrics.py"]Then navigate to the top-left application pane again and click “File” followed by “Save As.”
Name the text file “Dockerfile” (no extensions!) and save to your current working directory. VSC will auto-interpret the Docker language and you should notice a color change in the commands (FROM, RUN, COPY etc.).
In this Dockerfile, we’re using uv to install dependencies from a small, environment file called module3_reqs.txt.
Aside: When trying this yourself, perhaps you’re fine with always just running the latest versions of anndata and scanpy, in which case, RUN pip install uv && uv pip install --system scanpy anndata works just as well.
Importantly, WORKDIR sets the working directory inside the container to /app. This directory exists independently of your host machine; it’s a path inside the Linux filesystem that the container sees. We’ll mount our data to /app in the next section.
Build Docker image and run container
Now we’ve got our .py script and our Dockerfile. Importantly, we’ve told Docker to add both the script and module3_reqs.txt to our image during the build by using the COPY command.
Now we can build the image and run the container. To first build the image, in your VSC integrated terminal (while in your working directory; or in /module3 if you’re following the associated repo), run the following:
docker build -t scanpy-qc .docker build
Tells Docker to build an image from the Dockerfile-t scanpy-qc
The-tflag tags the image with a name i.e., scanpy-qc.means “use the current directory as the build context” (command will fail otherwise)
Once that’s done, we can pass our pbmc3k.h5ad as input to our script and run within the scanpy-qc container we just created:
mkdir data
docker run --rm -v $(pwd):/app scanpy-qc /app/pbmc3k.h5ad /app/data/output_qc.h5ad-v $(pwd):/app- mountsmodule3(i.e., host folder) to/appin the container/app/qc-metrics.pyQC script/app/pbmc3k.h5ad- input file/app/data/output_qc.h5ad- deposits output in module3/data
Now our output_qc.h5ad with QC metrics appended is in module3/data!
Here’s a summary visual of how the Dockerfile/image, container and host mounts interact:
[Host Machine]
├─ module3/
│ ├─ qc-metrics.py ← script (baked into image)
│ ├─ module3_reqs.txt ← env requirements (baked into image)
│ ├─ pbmc3k.h5ad ← input data
│ └─ data/ ← output folder
│ docker build
▼
[Docker Image: scanpy-qc]
├─ Python base OS (bullseye)
├─ Installed packages (uv, scanpy, anndata)
├─ qc-metrics.py ← copied in
└─ module3_reqs.txt ← copied in
│ docker run -v $(pwd):/app
▼
[Docker Container: running instance]
├─ /app/qc-metrics.py ← from image
├─ /app/module3_reqs.txt ← from image
├─ /app/pbmc3k.h5ad ← from host mount
└─ /app/data/ ← output folder, mapped to host
The power of containerization lies in the fact that we can make this Docker image easily accessible to our bioinformatics teammates for running our QC-metrics script on their machines with their own data, without needing to spend time versioning, fighting with dependencies or setting up environments.
To do so, let’s save our image to a tarball in our host /module3 directory:
docker save -o scanpy-qc.tar scanpy-qc(Note: You won’t see scanpy-qc.tar in the repo because I’ve excluded .tar files from being tracked via .gitignore. Originally, trying to push the image broke git because it was too large).
Now, all our teammates need to run in order to use the image is:
docker load -i scanpy-qc.tarThis imports the image to their local Docker. Now they only need to create /data in their working directory of choice, ensure that their actual data is in their top-level directory and run the exact same docker run command we did above. Their output file will be deposited just the same!
Conclusion
Using software like Docker for containerization provides a reproducible and shareable environment for computational analyses. In commercial R&D, this ensures that complex, commonly run workflows for single-cell analysis like QC annotation run consistently across different machines, teams, or even geographic locations without the, “it works on my machine” challenge.
Containers are “self-contained,” encapsulating all dependencies, libraries and scripts, eliminating the need for colleagues to manually install and configure dozens of packages. As you might imagine this greatly reduces setup time, prevents version conflicts and minimizes human error.
For a colleague to run a containerized analysis, they’d only need:
The container image (provided as a .tar, as we’ve done)
Input data in a path accessible to the container
The command to run the container (i.e.,
docker run --rm -v $(pwd):/app scanpy-qc /app/pbmc3k.h5ad /app/data/(exactly as we’ve done above)
In our next lesson, we’ll take reproducibility a step further and talk about scale. Our colleague can now run our analysis exactly, but what happens when our data only contained 3,000 samples, while theirs span 300,000?
In the meantime, if you’d like to play around with other Docker images specifically for single-cell analysis, these two are solid: one for inferring cell-cell communication from scRNA-seq data, and another larger, more general image for sc-analysis. Feel free to browse the Docker Hub image library for more.
All code available here.


Aneesa, thank you so much.
I will make sure to follow this step to ensure my codes are reproducible.
I will be sending my questions from previous modules till now to you soonest.
Once again, thank you for all you are doing.